Emerging targets in cancer drug resistance
Abstract
Drug resistance is a complex phenomenon that frequently develops as a failure to chemotherapy during cancer treatment. Malignant cells increasingly generate resistance to various chemotherapeutic drugs through distinct mechanisms and pathways. Understanding the molecular mechanisms involved in drug resistance remains an important area of research for identification of precise targets and drug discovery to improve therapeutic outcomes. This review highlights the role of some recent emerging targets and pathways which play critical role in driving drug resistance.
Keywords
Introduction
Drug resistance during cancer treatment frequently originates with the failure of chemotherapy. The term “chemotherapy” was first introduced 70 years back by Goodman and co-workers for the treatment of leukemia and lymphosarcoma at the end of the II World war[1]. Since then chemotherapy remains a mainstay treatment modality in cancer management. A large number of targets and treatment approaches to cancer have recently emerged, however pertinent resistance and severe side-effects remains a major clinical problem. A large patient inter-individual variability in their pharmacokinetics and inconsistent antitumor effects has been observed for most anticancer drugs[2]. However, most patients do not respond to these drugs and they often experience severe adverse effects such as neutropenia, diarrhea and potential hair loss. The major reason for this effect remains indiscriminate targeting of both normal and malignant cells by these chemotherapeutic agents. More than 200 anticancer drugs including cytotoxic and biologically targeted agents are currently in use having a low success rate of ~5% in the clinical application[3]. Therefore, identification of novel drug targets and development of effective chemotherapeutic agents to overcome drug resistance in cancer remains a major priority. Based on tumor response to the initial therapy, chemo-resistance mechanisms are classified into two categories including de novo (intrinsic) and acquired (extrinsic), however, the detailed mechanism(s) of chemo-resistance in human cancers remains to be understood completely.
Previous studies have identified drug efflux transporters/multi drug resistance pumps, uridine diphospho-glucuronosyltransferase (UGT) superfamily, cytochrome P450s and Glutathione S-transferases (GSTs) as some classical drug resistance target(s) in cancer chemotherapy[4-6]. GSTs includes eukaryotic and prokaryotic phase II family isozyme. For the detoxification process, GST catalyzes the reduced form of glutathione (GSH) to xenobiotic substrates. GST expression levels are linked with higher tumor drug resistance. Another GST specific isoenzyme called Glutathione S-transferases P are ubiquitous in nature and showed elevated levels in non-drug resistant and drug-resistant cancers[7]. ATP-binding cassette (ABC) transporters are the membrane-bound proteins require ATP for functioning. These transporters allow the substrates in (influx) or out (efflux) of the cells. Higher expression of ABC transporter also results in efflux of cytotoxic agent from the cancer cells which leads to drug resistance. In addition to above mechanisms of drug resistance, mutation-induced alterations in cytochrome P450 activity or other types of modifications in malignant cells including glycosylation of anticancer agents by UGT superfamily may modify drug efficacy as a result of altered drug metabolism[8].
More recent studies have identified abrupt cellular signaling pathways as underlying mechanisms for the development of cancer drug resistance. Some key process involve in drug resistance include tumor heterogeneity, reactivation of drug targets, hyperactivation of alternative pathways, cross-talk with the microenvironment, altered DNA response and its repair, modification in epigenetic pathways, impairment in apoptosis/autophagy and existence of cancer stem cells[9]. Any alteration in these pathways initially initiates changes in metabolic pathways and alteration in endocrine system functioning with later association with uncontrolled cell division through gene regulation. Innovations in molecular and biochemical techniques led to the identification/establishment of some new pathways including transforming growth factor beta (TGF-β) and Keap1-Nrf2 and some emerging drug resistance targets such as FOXO3A, FOXM1, FAKs, ANXA2, KCNN3, MIEN1 and epigenetic modifiers such as miRNAs and alternative splicing. In the present review we discussed these novel anticancer drug resistant targets and pathways.
Drug transport and metabolism
Various trans-membranous proteins such as ABC have been well reported for the resistance initiation against several chemotherapeutic drugs. These include P-glycoprotein known as multi-drug resistance protein 1 (MDR1), ABCC1 known as MDR-associated protein 1 (MRP1) and ABCG2 known as breast cancer resistance protein (BCRP). These transporter proteins are highly specific to eliminate cancer therapeutic drugs such as topoisomerase inhibitors, taxanes, and antimetabolites. Epithelial cells which participate in the excretion process express higher levels of MDR1 proteins[10]. Several studies demonstrate that overexpression of MDR1 in different cancers such as colorectal, hepatocellular, renal, breast, lung, prostate, lymphomas and leukemia’s have been tightly associated with chemo-resistance[11-14]. Breast cancer resistance protein also possess chemo-resistive properties against leukemia[15,16]. Some cancer therapeutic drugs such as nilotinib, erlotinib, sunitinib, and imatinib are prime target for MDR1 and BCRP efflux pumps[17]. MDR1 pump inhibitor, tariquidar in combination with anthracycline/taxanes showed partial activity in stage III/IV breast carcinoma during clinical trials[18]. In addition to this, another MDR1 inhibitor zosuquidar in combination with docetaxel did not show any effect in overall survival[19]. These facts suggest that the ABC transporter family comprise a high degree of functional redundancy.
Metabolism of chemotherapeutic drugs also possesses a major role in drug resistance initiation. Several studies have shown that activation or inactivation of chemotherapeutic drugs leads to drug resistance. Meijer et al.[20] demonstrated that the thiol group of glutathione inactivates platinum-based chemotherapeutic drugs. Absence or inactive form of the cellular enzyme that converts methotrexate and 5-fluorouracil (5-FU) to their active forms are also responsible for drug resistance[21,22]. Thymidine phosphorylase converts fluoropyrimidine prodrug (capecitabine) to 5-FU[23]. Methylation of thymidine phosphorylase encoding gene can cause capecitabine resistance[24]. UDP glucuronosyltransferase 1 promoter methylation inactivates its expression which positively regulates topoisomerase I inhibitor irinotecan drug and protects it’s from deactivation[25,26].
Cellilar signaling pathways in drug resistance
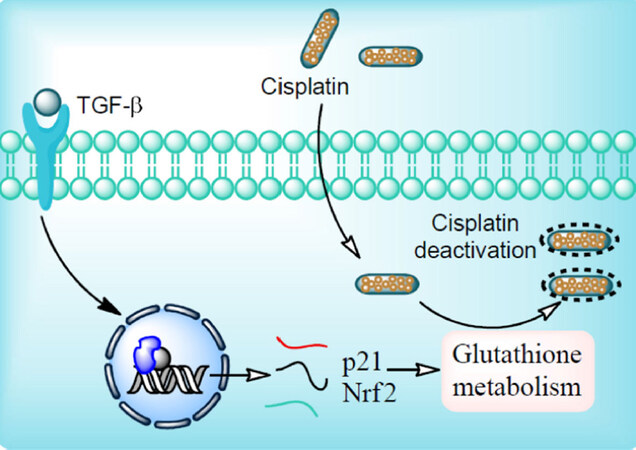
The TGF-β pathway regulates numerous cellular processes including cellular growth and differentiation, apoptosis and maintains cellular homeostasis[27]. TGF-β plays an important role in the maintenance of tumor microenvironment and its subsequent progression. Both intracellular and extracellular signals (DNA damage and TGF-β) have potential to induce p21/Waf1 gene transcription. Oxidative stress also elevates the p21/Waf1 and Nrf2 gene expression. Together, these proteins remarkably enhance the glutathione metabolism and reduce the potential of the anticancer drug such as cisplatin[28][Figure 1].
Figure 1. Mechanism of cisplatin resistance. Ligand binding to transforming growth factor beta (TGF-β) receptor initiates intracellular signaling through Smad protein complex (SPC). In the nucleus, SPC bind with the DNA binding domain which results in expression of p21/Waf1 and Nrf2 gene. p21/Waf1 and Nrf2 gene products tightly regulate glutathione metabolism. Cisplatin enters cells by passive diffusion. At low chloride ion concentration, the chloride ion of cisplatin is replaced with water molecules and forms activated cisplatin (aquation). Activated cisplatin enters the nucleus and results into the transcription of genes involved in anticancer activity. Glutathione conjugation with cisplatin hinders its nuclear translocation and thereby its chemo-preventive potential resulting into cisplatin resistance
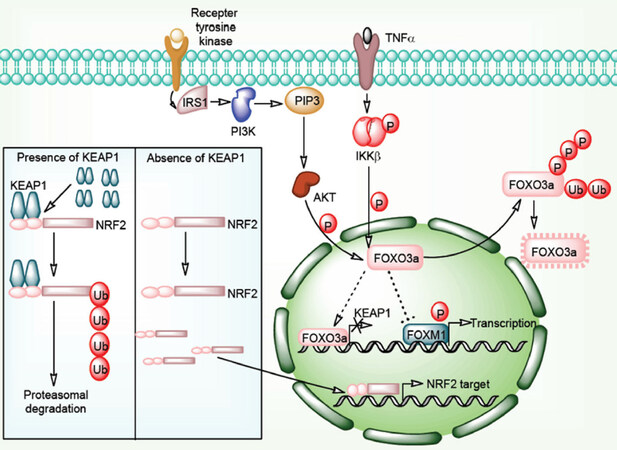
The Cap’n’collar (CNC) family proteins play important role in gene transcription and mammalian developmental process. Nuclear factor erythroid 2-related factor 2 (Nrf2) is an important member of CNC family proteins[29]. Nrf2 is a transcription factor known to modulate the expression of genes involved in the cellular antioxidant-oxidant system. Kelch-like ECH-associated protein 1 (Keap1) is an E3 ubiquitn ligase adaptor protein responsible for its interaction with the Nrf2 protein. Double glycine repeat (DGR) and c-terminal region of Keap1 interacts with the motifs of Nrf2. This interaction results in the proteasomal degradation of Nrf2 transcription factor in a constitutive manner. Oxidative stress weakens the Keap1-Nrf2 interaction resulting into the dissociation and translocation of Nrf2 into the nucleus. In the nucleus Nrf2 induces the transcription of genes involved in cytoprotection and metabolic pathways. In malignant cells, constitutive degradation of Nrf2 is disrupted which results in the increase in Nrf2 responsive gene expression. Keap1 and Nrf2 gene mutations, exon loss in Nrf2 gene, methylation of Keap1 promoter, sequestosome 1 protein accumulation, fumarate hydratase mutation, and activation of Nrf2 gene are some critical factors involved in the Nrf2 induction[30][Figure 2].
Figure 2. Keap1-Nrf2 signaling pathway in cancer drug resistance. Interaction of Keap1 molecules to Nrf2 protein is followed by Cul3-based E3 ligase complex mediated Nrf2 polyubiquitylation results into its proteasomal degradation. In the absence of Keap1 molecules, Nrf2 freely enters in the nucleus and transcribes its target genes in association with other nuclear factors. PI3K/Akt/TNF-α/NF-κB pathway directly phosphorylates FOXO3a proteins and directs them for ubiquitination and proteasomal degradation. Normally, FOXO3a proteins inhibits FOXM1 function and represses FOXM1 targeted transcription. FOXO3a protein also transcribes Keap1 genes. Absence of FOXO3a protein results in to downregulation of Keap1mRNA and FOXM1 targeted genes
Apart from the TGF-β and Keap1-Nrf2 signaling, mutation in the drug transporter gene could lead to drug resistance. Cisplatin uptake transporter named as CTR1 (SLC31A1) can be regulated by mutations which result in the introduction of drug resistance[31]. Mutation in the reduced folate carrier (RFC1/SLC19A1) protein causes methotrexate (MTX) drug resistance[32]. This could be achieved by efflux of intracellular MTX by MRP1 drug efflux pumps. Alteration in the MTX polyglutamylation also diminishes the MTX cellular sensitivity. MTX polyglutamates can also prevent dihydrofolate reductase (DHFR) enzyme activity inhibiting the conversion of dihydrofolate (FH2) to tetrahydrofolate (FH4)[33]. Absence of tetrahydrofolate reduces DNA synthesis and induces cell death. Mutation in DHFR enzyme induces obstruction between the interaction of MTX and its polyglutamates. MTX and its polyglutamates suppresses thymidylate synthesis which also diminishes DNA synthesis. Alterations in the levels or affinity of these enzymes in the cellular system develop the drug resistance. Drug resistance can also be developed by high copy number of the DHFR gene expression[34].
Inside the cell, water replaces the Cl groups of cisplatin and generates reactive nucleophilic species. These species have the ability to penetrate the nuclear membrane and can form covalent bond with the DNA molecule (primarily guanine) which initiates the process of cell death. Activation of nucleotide excision repair or other repair pathway introduces resistance in cancer cells. Prior to cisplatin entering in the nucleus, glutathione conjugation with cisplatin by the enzyme GSH S-transferases reduces drug effectiveness[35]. Cisplatin interaction with the sulfhydryl-containing metallothioneins also diminishes drug efficacy. Alternative pathway of cellular cisplatin resistance includes the efflux activity of the MRP2 (ABCC2) transporter and ATP7B P-type adenosine triphosphatase (ATPase) transporter[36].
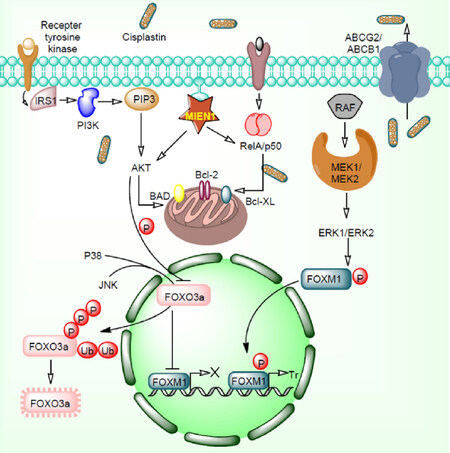
The EGFR/PI3K-PTEN/Akt/mTOR/ and Wnt/β-catenin signaling pathways play important roles in the malignant transformation and drug resistance[37]. These signaling pathways are frequently altered in various cancers due to the anomalous expression levels of PTEN, HER2, EGFR1, and other tumor suppressor gene/oncogenes. Some of the kinases involved in the cellular signaling also play a comprehensive role in cancer development and drug resistance establishment[38]. Due to its substantial implication in drug resistance development, kinases are the second largest drug target family. In addition to this, tumor microenvironment positively regulates these kinases and helps tumor to acquire resistance properties. Several signaling pathways have been reported for their outstanding contribution to the maintenance of tumor microenvironment. Mutation in EGFR and its downstream signaling leads to the expansion of cancer cell population[39]. Another transcription factor, polyoma enhancer activator 3 (Pea3) has been reported for its pivotal engrossment in invasion and metastasis in small cell lung cancer. Vascular endothelial growth factor stimulates angiogenesis and vasculogenesis (blood vessel formation) having potential to induce drug resistance. Another well-known signaling molecule Wnt16B regulates important factor required for cell proliferation and epithelial-mesenchymal transition in cancer cells[40]. Nuclear factor-κB (NF-κB) regulates the Wnt16B expression levels during these events. These pathways create a boundary and limit the drug efficacy and lead to development of drug resistance. Additionally, malignant cells secrete plasminogen activator inhibitor-1 which provides aggressiveness and facilitates cell proliferation[41]. AKT and ERK1/2 signaling and reduced level of caspase 3 participate in these events [Figure 3].
Figure 3. EGFR/PI3K/MAPK/ERK1/2-FOXO3a pathways in cancer drug resistance. Overexpression of MIEN1 and ABCG2/ABCB1 initiates the cisplatin resistance by targeting Akt/RelA/p50 and efflux of cisplatin respectively. MIEN1 targets Akt/RelA/p50 and induces overexpression of the anti-apoptotic proteins. Akt protein activated by MIEN1 inhibits FOXO3a function. Together, P38 and JNK phosphorylate FOXO3a protein which results in to its ubiquitination and proteasomal degradation. MEK/ERK pathway phosphorylates FOXM1 proteins which results in to translocation of these proteins inside the nucleus. Phosphorylated FOXM1 transcribes several genes which positively involves in drug resistance
Genetic modifications in drug resistance
DNA mutation
Variation in expression or mutation in the drug target proteins may be the major reason for drug resistance initiation. Fluorodeoxyuridine monophosphate (FdUMP) actively targets thymidyate synthase[42]. One of the studies demonstrated that thymidyate synthase expression determines the 5-FU sensitivity[43]. Thymidyate synthase promoter polymorphism (TSER3/TSER3) negatively regulates 5-FU-based chemotherapy than the heterozygous (TSER2/TSER3) /homozygous alternative polymorphism (TSER2/TSER2)[44]. Overexpression of thymidyate synthase encourages resistance to its inhibitors like TDX, 5-FU, and multi-targeted anti-folate[45,46]. SN-38, a metabolite of CPT-11, inhibits DNA topoisomerase-I enzyme which relaxes super-coiled double-stranded DNA during the replication process[47]. Downregulation of topoisomerase-I mRNA results in anti-sensitivity against CPT-11 in colorectal cancer[48]. Anthracyclines (doxorubicin) and epipodophyllotoxins (etoposide) are ideal target for the topoisomerase-II enzyme which also plays a vital role during replication. Alteration in topoisomerase-II expression levels or mutation in this protein results in resistant to doxorubicin/etoposide in colon cancer cells[49,50].
Tubulin proteins such as α- and β-tubulin generate microtubule structures that maintain cell integrity, regulates signaling/cell division and helps in vesicle transportation throughout the cellular compartments[51]. Tubulin polymerization/de-polymerization regulates appropriate spindle formation and chromosome segregation during cell division. Taxanes (paclitaxel and docetaxel) and vinca alkaloids (vinblastine and vincristine) repress the polymerization of tubulin which blocks cell division[52,53]. An in vitro study reported that mutation in the β-tubulin proteins regulates the sensitivity against paclitaxel[54,55].
DNA repair
Mutation in gene encoding enzymes involved in DNA damage response (DDR) such as ataxia telangiectasia (ATM), ATR, RAD50, and WRN can enhance the risk of cancer emergence and resistance[56]. Single nucleotide polymorphisms in DNA repair-associated genes can also induce cancer drug resistance[57]. Some of the chemotherapeutic drugs such as 5-fluorouracil, epirubicin, cisplatin, and doxorubicin have been well studied in cancer for their potential to cause damage in the DNA molecule. DNA damage triggers DDR proteins to repair the DNA and induces drug resistance in cancer cells[58]. DNA repair associated (DRCA) genes directly regulate DNA repair pathway via the homologous recombination and DNA strand breaks. Development of inhibitors for these proteins are directly involved in the repair pathway can reduce drug resistance and sensitize to these agents. Poly ADP-ribose polymerase 1 (PARP1) is an important protein which modified nuclear proteins through the poly ADP-ribosylation and makes them active for the DNA damage repair. Olaparib, USFDA approved drug, is a PARP inhibitor which blocks DNA repair functions and induces apoptosis in cancer cells[58]. Another DNA repair gene namely NEK2, codes serine/threonine mitotic kinase plays an important role in spindle formation and chromosome segregation[59,60]. Silencing of NEK2 reduces drug resistance to bortezomib in vitro and in vivo. NER pathway comprises excision repair cross-complementing 1, whose increased levels has been linked to poor response to chemotherapy in ovarian, gastric and non-small-cell lung carcinoma[61,62]. Cisplatin-treated testicular cancers showed decreased levels of excision repair cross-complementing 1[63].
Epigenetic modifications in drug resistance
Alternative splicing
Splicing events after the post transcription process matures the mRNA to become respective functional protein. Any deformities in these processes produce nonfunctional or mutated proteins resulting in cancer initiation and development. Alternative splicing accomplishes coding region reshuffling and regulates mRNA maturation[64]. Defects in splicing mechanism lead to drug resistance against selected anticancer agents. BIM, a type of BH3-only proteins belongs to the pro-apoptotic B-cell CLL/lymphoma 2 (BCL-2) protein family. It play critical role in hematopoietic homeostasis, autoimmune disease restriction and cancer initiation. BimEL, BimL and BimS are the three major BIM alternative splicing products[65]. Studies provide evidence that BIM alternative splicing molecules play a key role in drug resistance[66]. Upregulated BIM proteins facilitate tyrosine kinase inhibitors stimulated cell death events in kinase dependent tumors. Polymorphism also play a critical role in the origination of drug resistance. For example, an intron comprising 2903-bp deletion in BIM gene recruits privileged splicing of exon 3 over exon 4 produces BIM isoforms which does not bears BCL-2 homology domain 3 (BH3) domains. This type of BIM protein promotes drug resistance in NSCLC (non-small cell lung cancer) and CML (chronic myeloid leukemia) against tyrosine kinase inhibitors such as gefitinib and imatinib[67].
Folylpolyglutamate synthase (FPGS) enzyme is involved in the maintenance of folate homeostasis in the cytosolic and mitochondrial compartment. Exon 12 skipping in FPGS gene results in dysfunctional FPGS which leads to the inhibition of anti-folate drugs such as methotrexate and introduces resistance in acute lymphoblastic leukemia[68].
CART-19 immunotherapy is well known to recognize the full length CD19 protein which diminishes the death probability of cancer cells. Skipping of exon 2, in the maturation of CD19 mRNA, results in the truncated CD19 protein. This splicing event reduces the probability of tumor aggressiveness identification at the early stage and induces drug resistance[69]. In BRCA1 gene, skipping of exon 11 restores BRCA1 DNA repair potential and stimulates drug resistance[70].
DNA modification
Epigenetic modifications play a significant role in the expression of various receptor proteins such as hormone receptors. Tamoxifen treatment downregulates estrogen receptor expression after treatment in estrogen-responsive breast cancer. This may occur due to epigenetic silencing of the estrogen receptor-encoding genes. A study has shown that aberrant methylation of CpG islands of the estrogen receptor promoter and histone deacetylation downregulates its expression and induces tamoxifen resistance. Overexpression of HER2/EGFR, hypoxia and MAPKs hyper-activation downregulates the estrogen receptor expression. MiRNA alteration has been directly associated with tamoxifen resistance in breast cancer[71]. Steroid receptor coactivator-3 (SRC-3) regulates various processes of cancer development and act as a coactivator/transcription factor. Breast cancer cells have been reported for the overexpression of SRC-3 and positively correlated with tamoxifen resistance[72]. SRC-3 over-expression is also positively correlated with increased levels of HER-2/neu, poor disease-free survival and tamoxifen resistance in tamoxifen-treated breast cancer cells[73]. Epigenetic-driven drug resistance includes epigenetic modification in the gene of pro-apoptotic (DAPK, APAF-1), drug transporters (ABCB1), histone modifiers (KDM5A) and DNA repair proteins (MLH1, MGMT, FANCF). Chemotherapeutic drug treatment in drug-resistant cancer cells alters various enzymes (involved in genome regulation) which makes drug-resistant epigenome to drug-sensitive epigenome thereby rendering cancer cells sensitive to therapeutic drugs.
MicroRNA
Small noncoding RNA have been reported to control cell activity by regulating large number of target genes[74]. Recently researchers are focusing to acquire miRNA beneficial function with an attempt to overcome multidrug resistance-related phenomenon. MicroRNAs (miRNAs) are about 22-nucleotide long RNA produced from the proper processing of RNA structure. MiRNAs play a significant role in the regulation of gene expression[75]. It instructs numerous protein-coding genes including genes involved in cancer as well as drug resistance. Gene silencing involves the destruction of messenger RNA strand (mRNA) into two separate fragments. Gene silencing can also be achieved by diminishing the mRNA stability via shortening of poly (A) tail[76]. Another silencing mechanism included the reduced translation through ribosome of mRNA into proteins. Recent investigations have demonstrated that microRNAs play an important role in the development of drug resistance [Table 1]. MicroRNAs can assist as a biomarker for the patient survival in respect to drug-resistant therapies.
miRNAs involved in chemo-resistance in various cancer and their targets
| Chemotherapy agent | Target | Tumor | miRNAs | Ref. |
|---|---|---|---|---|
| Anthracyclines | MDR1 | SCLC | miR-7 | [77] |
| Temozolomide | MDR1/ABCG2 | Glioblastoma | miR-9 | [78] |
| Paclitaxel | PTEN,
p-gp/ABCB1 | Ovary, NSCLC | miR-17-5p, miR-145, miR-181a | [79-81] |
| Trastuzumab | PTEN, PDCD4 | Breast | miR-21 | [82] |
| Epriubicin | ABCG2 | Breast | miR-25 | [83] |
| Doxorubicin | P-gp,
MRP1/ABCC1 | Gastric, Brest | miR-103/107,
miR-134 | [84,85] |
| Adriamycin | MDR1/MRP1 | Glioma | miR-127 | [86] |
| Vincristine/cisplatin | ABCB1 | Gastric | miR-129-5p | [87] |
| 5-fluorouracil | ABCB1,
P-gp/ABCB1, ABCG2 | Gastric,
Colorectal | miR-129-5p,
miR-508-5p, miR-519c | [87-89] |
| Cisplatin | PTEN,
MDR1/MRP1, Cyclin D1, GRB2, ERK2, RSK1, RSK2 | NSCLC,
Ovary | miR-181a,
miR-196a, miR-634 | [81,90,91] |
| 5-fluorouracilmitomycin C | P-gp/ABCB1 | Colorectal | miR-200c | [92] |
| Vincristine/oxaliplatin/cisplatin | P-gp/ABCB1 | Colorectal,
Gastric | miR-200c,
miR-508-5p | [88,92] |
| Bortezomib | BAFF | Multiple myeloma | miR-202 | [93] |
| Thalidomide | BAFF | Multiple myeloma | miR-202 | [93] |
| Dexamethasone | BAFF | Multiple myeloma | miR-202 | [93] |
| Tamoxifen | PTEN | Breast | miR-217 | [94] |
| Lapatinib | PTEN | Breast | miR-217 | [94] |
| Etoposide | PTEN | Breast | miR-217 | [94] |
| Melphalan | MRP1/ABCC1 | Multiple myeloma | miR-221/222 | [95] |
| EGFR inhibitors | KRAS, AKT1 | NSCLC | [96] |
Novel targets in drug resistance
FOXO3a and FOXM1
Numerous cytosolic, nuclear, intracellular and extracellular factors have been reported in cancer development and drug resistance[97]. These factors directly/indirectly regulate several others factors and cellular signaling pathways. Some factors have repressive or stimulatory potential to each other resulting into therapeutic drug resistance and increased aggressiveness in cancer cells. Transcription factors superfamily comprising DNA binding domain includes Forkhead box (FOX) proteins. FOX has been reported to regulate cellular machinery and various homeostatic processes. FOX factors controls numerous physiological processes including cell division, cell death, cell invasion and migration and drug resistance[98-100]. FOXO3a receives signals from the signaling pathways such as EGFR/PI3K/Akt/ERK, and transfers it to other pathway which ultimately controls transcription of respective gene. Sometimes these factors alone bind to the regulatory region of the promoter and thereby regulate gene transcription. Hyperactivation of PI3K/Akt signaling inactivates FOX factors and induces drug resistance. Mutation in the PTEN gene and receptor tyrosine kinases overexpression together endorses cancer development and drug resistance[101]. Several studies revealed that anticancer drugs such as imatinib, tamoxifen, cisplatin, doxorubicin, paclitaxel, lapatinib and gefitinib induce FOXO activation via alteration in PI3K/Akt signaling pathways[99,102-105]. JNK signaling also promotes FOXO3a nuclear localization and activity by diminishing Akt phosphorylation[102,103][Figure 3]. The p38 protein actively phosphorylates FOXO3a at Serine 7 residue which in turn enhances its nuclear re-localization and provides doxorubicin directed response[98]. Another FOX family factor, FOXM1 acts like an oncogenic transcription factor mainly controls cell cycle and division. FOXM1 is overexpressed in several cancers including breast, liver, colorectal, lung and the prostate[106]. Oncogenic potential of FOXM1 and its uncontrolled cell division properties makes it available in the stem cell compartments and initiate hyperplasia. FOXM1 imparts resistance against chemotherapeutic drugs such as epirubicin and cisplatin [Figure 3]. It also induces drug resistance via overexpression of DDR gene viz. replication factor C4 (RFC4), epsilon 2, accessory subunit (POLE2), polo-like kinase 4 (PLK4), X-ray repair complementing defective repair in Chinese hamster cells (XRCC1), breast cancer type 2 susceptibility protein (BRCA2), polymerase (DNA directed) and exonuclease 1 (EXO1)[107-109]. Furthermore, FOXM1 and FOXO3a together compete for the same DNA sequences for binding. These factors also share numerous target genes which facilitates FOXM1 transcriptional output repression by FOXO3a proteins. However, FOXO3a prevents VEGF expression and FOXM1 facilitates VEGF overexpression to regulate cell migration, invasion and drug resistance.
Focal adhesion kinase
Focal adhesion kinase (FAK), a tyrosine kinase also referred as PTK2 (non-receptor protein tyrosine kinase) is a downstream protein of integrin and growth factor receptors signaling pathways. Increased expression of FAK has been associated with various type of cancer[110]. FAK modulates tumor progression, apoptosis, invasion, and metastasis. The N and C terminal domains (FERM and FAT domains) of the protein hinders the ATP binding site of the central kinase domain. The interaction between N and C terminal domain of the protein keeps it in an inactive form. Integrin and growth factor receptor activation induce disruption of N and C terminal domain resulting in the exposed ATP binding site on the central kinase domain. Binding of ATP to the central domain, phosphorylation of FAK-Y397 amino acid followed by SRC binding, and additional amino acid phosphorylation results in the activated FAK[111]. FAK induces PIP2/3 and AKT1-mediated survival signals in cells. FAK induces BCAR1 (breast cancer anti-estrogen resistance 1) and MAPK8 mediated cellular motility, proliferation, and survival in breast cancer. Beside FAK induces assembly and disassembly (turnover) of the focal adhesions and thus modulate migration of cells[112]. FAK is known to interact with some other proteins such as GRB2, TP53, MDM2, and RIP associated with the pathogenesis of cancer[113]. From the above discussion, it is clear that ATP binding domain and some other FAK domains involved in protein-protein interaction might serve as a cancer drug target. Inhibitors (VS-4718; VS-6062 and PF-573,228) having the binding ability to these domains serve as a promising candidate in anticancer drug discovery[113]. Studies indicate the promising efficacy of FAK inhibitors and chemotherapy synergism to reduce treatment side effects and drug resistance in cancer. These inhibitors increase chemosensitivity in drug-resistant cells and also synergize the drug treatment efficacy in cancer cells[113]. Recent reports suggest that FAK induces NF-κB pathway mediated cytokine production in response to DNA damage. This phenomenon protects the cells from DNA damage and maintains chemo-resistance in the cells[114]. In this regard, FAK inhibitors might play a critical role against DNA damage-mediated drug resistance in cancer cells.
ANXA2
Annexins are multifunctional proteins having ability to bind phospholipids in the presence of calcium ions. It plays important role in cytoskeleton dynamics, signal transduction, and membrane trafficking. Due to these associated phenomenon annexins are involved in the pathophysiological condition of various diseases including cancer[115]. Annexin 2 has annexin core structure (highly conserved), N terminal domain (highly variable), calcium-dependent lipid binding region, RNA-binding helices, F-actin binding site and C terminal[116]. Phosphorylation of annexin 2 protein is essential for its multi-functions. In response to cell transformation, Src kinase phosphorylates annexin 2 (45 phosphorylation sites are known) mainly at the N terminal domain or the central domain[116]. Phosphorylation of Tyr23 residue in annexin 2 proteins has been associated with tumor cell adhesion, angiogenesis, invasion, motility, progression, and proliferation[117]. Annexin 2, Tyr23 phosphorylation mediated STAT3 phosphorylation is known to involve in glucocorticoid resistance in cancer cells[118]. Annexin 2 induce coilin disruption/chromosome instability mediated cellular chemo-resistance in cancer cells[119]. ANXA2 and tenascin-C protein interaction mediated activation of PI3K/Akt/NF-κB signaling pathway is involved in drug resistance in prostate cancer[119]. Moreover, annexin 2 is also known to involve in radiotherapy and immunotherapy in different cancer cells.
Potassium calcium-activated channel subfamily N member 3
A potassium channel, potassium calcium-activated channel subfamily N member 3 (KCNN3) belongs to a trivial Ca2+-activated potassium channel family. KCNN3 have been reported for its role in solid tumor progression[120]. KCNN3 regulates cell membrane potential in melanoma and breast cancer cells[120]. Cell migration and invasiveness also partially regulated by P2X purinoceptor 7 and KCNN3. Transient receptor potential channel 1 and calcium release-activated calcium channel protein 1 with KCNN3 regulates store-operated calcium entry (SOCE)-dependent cell migration[121]. Upregulation of KCNN3 is positively associated with bortezomib-resistant myeloma cells which induce drug resistance[120].
Migration and invasion enhancer 1
Migration and invasion enhancer 1 (MIEN1) gene also known as C35, is a novel gene situated subsequent to the cluster of differentiation 340 (CD340) in the 17q12 amplicon of the humanoid chromosome[122]. The gene has unusual potential to increase tumor cell migration and invasion. MIEN1 expresses differentially in normal cells and cancer cells[123]. Regulation of MIEN1 by miRNAs may permit better targeting strategies to overcome the respective ailment. The hsa-miR-940 (miR-940) has been reported for their high expression levels in commemorated normal cells compared to tumor cells[123]. A study reported that hsa-miR-940 directly targets MIEN1 RNA and alters its downstream effectors in prostate cancer[123]. Inactivation of MIEN1 by miR-940 inhibits cell migration and invasion, diminished cell anchorage-independent growth aptitude and initiates overexpression of E-cadherin molecule which overcomes mesenchymal-to-epithelial transition (MET) process[123]. Another study reported that MIEN1, potassium calcium-activated channel subfamily N member 3 (KCNN3), and drug resistance in ovarian cancer are significantly associated with each other[120]. MIEN1 and DNp73 interaction induces chemo-resistance in ovarian cancer[124]. High levels of MIEN1 significantly promotes phosphorylation of tyrosine 23 (Y23) residue on annexin A2 (ANXA2) which facilitates the interaction with cellular actin filaments and regulates cell trickling/cytoskeletal rearrangement through actin transformation[119]. Earlier, it has been reported that ANXA2 expression correlates with poor prognosis and a higher number of chemotherapy cycles which stimulate chemoresistance[125]. Hypoxia and chemokines facilitate the transformation of human neural stem cells to glioma stem cells in presence of MIEN1[126]. FAK activating protein MIEN1 phosphorylates FAK at Y925 and induced phosphorylation of ERK1/2, Akt and NF-κB[127][Figure 3]. Previously it has been shown that FAK phosphorylation promotes overall survival of ovarian cancer patients[128]. MET process also promotes FAK phosphorylation and enhances taxane resistance in ovarian cancer[128]. High-mobility group box 2 (HMGB2) binding protein interacts with MIEN1 and enhances cancer progression and proliferation[129]. Silencing of HMGB2, an interacting partner MIEN1 sensitizes head and neck squamous cell carcinoma against cisplatin and 5-fluorouracil[130]. MIEN1 showed increased expression in lapatinib-sensitive breast cancer cells compared to lapatinib-resistant breast cancer cells[131]. Colony growth in soft agar, invasion into collagen matrix and formation of large acinar structures in three-dimensional cell cultures experiment demonstrated that increased MIEN1 expression is highly associated with cell transformation including epithelial to mesenchymal transition and reduced expression of E-cadherin and keratin-8[132]. This study also demonstrated that cell transformation was dependent on Syk kinases.
Sphingolipids
Membrane lipids family comprises sphingolipids, a fatty acid derivative of sphingosine which constructs lipid bilayer structure and maintains their fluidity[133]. Sphingolipids includes sphingosine 1-phosphate (S1P), ceramide, glucosylceramide (GlcCer) and sphingosine that regulates various biological events such as proliferation, apoptosis, inflammation, senescence and cell migration in cancer cells[133,134]. Alteration of sphingolipid metabolism, as well as ceramide accumulation, is reported as a major factor for resistance development against chemotherapy in cancer cells[135]. Ceramide metabolism vastly produces GlcCer as a product due to higher glucosylceramide synthase (GCS) enzyme activity in tumor cells[136]. Studies have demonstrated that GCS overexpression and its activity, positively correlated with ABC transporter facilitating drug resistance[137,138]. GCS knockdown significantly reduces the MDR1 expression, a gene that encodes for ABC transporter protein[137,138]. ABC family transporters also transport sphingolipids, phospholipids, and glucosylceramide across the lipid bilayer.
Sphingosine kinase 1 (SK1) and S1P metabolism regulate drug sensitivity against cancer cells because overexpression of these molecules provides shelter to cancer cells from drug treatment. A study reported its increased levels in camptothecin resistant prostate cancer cells[139]. Another in vivo study demonstrated that SK1 and S1P inflection results in cisplatin sensitivity toward the cellular slime mold Dictyostelium discoideum, a powerful non-mammalian model for drug target discovery and resistance[140]. Baran et al.[141] reported that imbalance between C18-ceramide and S1P is associated with SK1 overexpression and BCR-ABL upregulation which ultimately leads to imatinib resistance in human chronic myeloid leukemia K562 cells. Downregulation of SK1 levels enhanced the imatinib drug sensitivity and induces the apoptosis in these cells. Another in vivo study demonstrated that silencing of sphingosine 1 phosphate phosphatase 1 through miR-95, endorses S1P-dependent resistance to radiation in breast/prostate tumors[142].
Future perspectives
Drug resistance in cancer, either intrinsic or acquired, substantially reduces the efficacy of chemotherapeutic drugs with poor prognosis in cancer patients. To achieve higher likelihood of therapeutic success, a complete understanding of the mechanisms underlying chemo-resistance is needed. Recent development of high-throughput screening technologies has enhanced the identification of intrinsic and extrinsic cellular pathways that may be targeted to prevent or reverse drug resistance. Other evolving techniques including open reading frame screens, RNA interference, genome editing, and proteomics analysis of drug resistant cell lines and tissues will provide important information and identification of novel targets to overcome tumor drug resistance. In addition, smarter means to deliver anticancer drugs through targeted nanotechnology approach is being tested. This knowledge will be extremely helpful for the development of precision therapies based on the prediction of tumor cell response to the currently available chemotherapeutic agents and also the discovery of novel therapeutic strategies to treat cancer or reverse tumor chemo-resistance. Further work is required to determine which subset of cancer patients are suitable candidates for a particular multi-targeted therapy or combination regimen affecting multiple targets. Additional research or modeling is also needed to identify what combination of targets can be expected to optimize therapy for particular cancer types. Such new knowledge will be translated into the development of innovative cancer therapeutics to overcome drug resistance.
Conclusion
Increased understanding of the mechanisms underlying cancer drug resistance suggests that an integrated approach to cancer therapy is needed for targeting multiple signaling pathways. Recent use of molecularly targeted agents to target multiple signaling pathways remains an important approach in cancer treatment, however, use of these targeted therapies is not without limitations. Further research is needed to identify approaches to repurpose drugs to optimize therapy for particular cancer types.
Declarations
Authors’ contributionsSubstantial contributions to conception and writing of the manuscript: Kumar S, Gupta S
Developing figures and tables: Kumar S, Kushwaha PP
Availability of data and materialsNot applicable.
Financial support and sponsorshipEfforts are supported by the Department of Defense Grants (W81XWH-18-1-0618, W81XWH-15-1-0558); VA Merit Review (1I01BX002494) to Gupta S. Kushwaha PP acknowledges financial support from University Grants Commission, India in the form of CSIR-UGC Senior Research fellowship. Kumar S acknowledges Department of Science and Technology, India, and University Grants Commission, India for providing financial support in the form of DST-SERB Grant [EEQ/2016/000350] and UGC-BSR Research Start-Up-Grant [No. F.30–372/2017 (BSR)] respectively.
Conflicts of interestAll authors declared that there are no conflicts of interest.
Ethical approval and consent to participateNot applicable.
Consent for publicationNot applicable.
Copyright© The Author(s) 2019.
REFERENCES
1. Goodman LS, Wintrobe MM, Dameshek W, Goodman MJ, McLennan MT, et al. Nitrogen mustard therapy: the use of methyl-bis(β-chloroethyl) amine hydrochloride and tris(\sb-chlorethyl) amine hydrochloride for Hodgkin’s disease, lymphosarcoma, leukemia, and certain allied and miscellaneous disorders. JAMA 1946;132:126-32.
2. Holohan C, Van Schaeybroeck S, Longley DB, Johnston PG. Cancer drug resistance: an evolving paradigm. Nat Rev Cancer 2013;13:714-26.
4. Glavinas H, Krajcsi P, Cserepes J, Sarkadi B. The role of ABC transporters in drug resistance, metabolism and toxicity. Curr Drug Deliv 2004;1:27-42.
5. McCubrey JA, Abrams SL, Fitzgerald TL, Cocco L, Martelli AM, et al. Roles of signaling pathways in drug resistance, cancer initiating cells and cancer progression and metastasis. Adv Biol Regul 2015;57:75-101.
6. Cazenave LA, Moscow JA, Myers CE, Cowan KH. Glutathione S--transferase and drug resistance. In: Robert FO, editor. Drug resistance in cancer therapy. Boston, MA: Springer; 1989. pp. 171-87.
7. Zhang J, Grek C, Ye ZW, Manevich Y, Tew KD, et al. Pleiotropic functions of glutathione S-transferase P. Adv Cancer Res 2014;122:143-75.
8. Islam MT. Hallmarks of cancer drug resistance and overcoming ways: a paradigm. Int J Pharm Biol Arch 2017;8:1-17.
10. Choi CH. ABC transporters as multidrug resistance mechanisms and the development of chemosensitizers for their reversal. Cancer Cell Int 2005;5:1-13.
11. Thomas H, Coley HM. Overcoming multidrug resistance in cancer: an update on the clinical strategy of inhibiting p-glycoprotein. Cancer Control 2003;10:159-65.
12. Triller N, Korošec P, Kern I, Košnik M, Debeljak A. Multidrug resistance in small cell lung cancer: expression of P-glycoprotein, multidrug resistance protein 1 and lung resistance protein in chemo-naive patients and in relapsed disease. Lung Cancer 2006;54:235-40.
13. Nooter K, De La Riviere GB, Look MP, Van Wingerden KE, Henzen-Logmans SC, et al. The prognostic significance of expression of the multidrug resistance-associated protein (MRP) in primary breast cancer. Br J Cancer 1997;76:486-93.
14. Zalcberg J, Hu XF, Slater A, Parisot J, El-Osta S, et al. MRP1 not MDR1 gene expression is the predominant mechanism of acquired multidrug resistance in two prostate carcinoma cell lines. Prostate Cancer Prostatic Dis 2000;3:66-75.
15. Doyle LA, Yang W, Abruzzo LV, Krogmann T, Gao Y, et al. A multidrug resistance transporter from human MCF-7 breast cancer cells. Proc Natl Acad Sci U S A 1998;95:15665-70.
16. Robey RW, Shukla S, Steadman K, Obrzut T, Finley EM, et al. Inhibition of ABCG2-mediated transport by protein kinase inhibitors with a bisindolylmaleimide or indolocarbazole structure. Mol Cancer Ther 2007;6:1877-85.
17. Shukla S, Chen ZS, Ambudkar SV. Tyrosine kinase inhibitors as modulators of ABC transporter-mediated drug resistance. Drug Resist Updat 2012;15:70-80.
18. Pusztai L, Wagner P, Ibrahim N, Rivera E, Theriault R, et al. Phase II study of tariquidar, a selective P-glycoprotein inhibitor, in patients with chemotherapy-resistant, advanced breast carcinoma. Cancer 2005;104:682-91.
19. Ruff P, Vorobiof DA, Jordaan JP, Demetriou GS, Moodley SD, et al. A randomized, placebo-controlled, double-blind phase 2 study of docetaxel compared to docetaxel plus zosuquidar (LY335979) in women with metastatic or locally recurrent breast cancer who have received one prior chemotherapy regimen. Cancer Chemother Pharmacol 2009;64:763-8.
20. Meijer C, Mulder NH, Timmer-Bosscha H, Sluiter WJ, Meersma GJ, et al. Relationship of cellular glutathione to the cytotoxicity and resistance of seven platinum compounds. Cancer Res 1992;52:6885-9.
21. Schwartz PM, Moir RD, Hyde CM, Turek PJ, Handschumacher RE. Role of uridine phosphorylase in the anabolism of 5-fluorouracil. Biochem Pharmacol 1985;34:3585-9.
22. Houghton JA, Houghton PJ. Elucidation of pathways of 5-fluorouracil metabolism in xenografts of human colorectal adenocarcinoma. Eur J Cancer Clin Oncol 1983;19:807-15.
23. Malet-Martino M, Martino R. Clinical studies of three oral prodrugs of 5-fluorouracil (capecitabine, UFT, S-1): a review. Oncologist 2002;7:288-323.
24. Kosuri KV, Wu X, Wang L, Villalona-Calero MA, Otterson GA. An epigenetic mechanism for capecitabine resistance in mesothelioma. Biochem Biophys Res Commun 2010;391:1465-70.
25. Belanger AS, Tojcic J, Harvey M, Guillemette C. Regulation of UGT1A1 and HNF1 transcription factor gene expression by DNA methylation in colon cancer cells. BMC Mol Biol 2010;11:1-11.
26. Toffoli G, Cecchin E, Gasparini G, D’Andrea M, Azzarello G, et al. Genotype-driven phase I study of irinotecan administered in combination with fluorouracil/leucovorin in patients with metastatic colorectal cancer. J Clin Oncol 2010;28:866-71.
28. Oshimori N, Oristian D, Fuchs E. TGF-β promotes heterogeneity and drug resistance in squamous cell carcinoma. Cell 2015;160:963-76.
29. Chan K, Lu R, Chang JC, Kan YW. NRF2, a member of the NFE2 family of transcription factors, is not essential for murine erythropoiesis, growth, and development. Proc Natl Acad Sci U S A 1996;93:13943-8.
30. Jung BJ, Yoo HS, Shin S, Park YJ, Jeon SM. Dysregulation of NRF2 in cancer: from molecular mechanisms to therapeutic opportunities. Biomol Ther 2018;26:57-68.
31. Ishida S, Lee J, Thiele DJ, Herskowitz I. Uptake of the anticancer drug cisplatin mediated by the copper transporter Ctr1 in yeast and mammals. Proc Natl Acad Sci U S A 2002;99:14298-302.
32. Yee SW, Gong L, Badagnani I, Giacomini KM, Klein TE, et al. SLC19A1 pharmacogenomics summary. Pharmacogenet Genomics 2010;20:708-15.
33. Hagner N, Joerger M. Cancer chemotherapy: targeting folic acid synthesis. Cancer Manag Res 2010;2:293-301.
34. Morales C, García MJ, Ribas M, Miró R, Muñoz M, et al. Dihydrofolate reductase amplification and sensitization to methotrexate of methotrexate-resistant colon cancer cells. Mol Cancer Ther 2009;8:424-32.
35. Barabas K, Milner R, Lurie D, Adin C. Cisplatin: a review of toxicities and therapeutic applications. Vet Comp Oncol 2008;6:1-8.
36. Surowiak P, Materna V, Kaplenko I, Spaczynski M, Dolinska-Krajewska B, et al. ABCC2 (MRP2, cMOAT) can be localized in the nuclear membrane of ovarian carcinomas and correlates with resistance to cisplatin and clinical outcome. Clin Cancer Res 2006;12:7149-58.
37. Davis NM, Sokolosky M, Stadelman K, Abrams SL, Libra M, et al. Deregulation of the EGFR/PI3K/PTEN/Akt/mTORC1 pathway in breast cancer: possibilities for therapeutic intervention. Oncotarget 2014;5:4603-50.
38. Barouch-Bentov R, Sauer K. Mechanisms of drug resistance in kinases. Expert Opin Investig Drugs 2011;20:153-208.
39. Bonavia R, Mukasa A, Narita Y, Sah DW, Vandenberg S, et al. Tumor heterogeneity is an active process maintained by a mutant EGFR-induced cytokine circuit in glioblastoma. Genes Dev 2010;24:1731-45.
40. Sun Y, Campisi J, Higano C, Beer TM, Porter P, et al. Treatment-induced damage to the tumor microenvironment promotes prostate cancer therapy resistance through WNT16B. Nat Med 2012;18:1359-68.
41. Kang J, Kim W, Kwon T, Youn H, Kim JS, et al. Plasminogen activator inhibitor-1 enhances radioresistance and aggressiveness of non-small cell lung cancer cells. Oncotarget 2016;7:23961-74.
42. Peters GJ, Kohne CH. Fluoropyrimidines as antifolate drugs. In Antifolate Drugs in Cancer Therapy. Totowa, NJ: Humana Press; 1999. pp. 101-45.
43. Longley DB, Harkin DP, Johnston PG. 5-fluorouracil: mechanisms of action and clinical strategies. Nat Rev Cancer 2003;3:330-8.
44. Marsh S, McLeod HL. Thymidylate synthase pharmacogenetics in colorectal cancer. Clin Colorectal Cancer 2001;1:175-8.
45. Longley DB, Ferguson PR, Boyer J, Latif T, Lynch M, et al. Characterization of a thymidylate synthase (TS)-inducible cell line: a model system for studying sensitivity to TS-and non-TS-targeted chemotherapies. Clin Cancer Res 2001;7:3533-9.
46. Longley DB, Boyer J, Allen WL, Latif T, Ferguson PR, et al. The role of thymidylate synthase induction in modulating p53-regulated gene expression in response to 5-fluorouracil and antifolates. Cancer Res 2002;62:2644-9.
47. Xu Y, Villalona-Calero MA. Irinotecan: mechanisms of tumor resistance and novel strategies for modulating its activity. Ann Oncol 2002;13:1841-51.
48. Boyer J, McLean EG, Aroori S, Wilson P, McCulla A, et al. Characterization of p53 wild-type and null isogenic colorectal cancer cell lines resistant to 5-fluorouracil, oxaliplatin, and irinotecan. Clin Cancer Res 2004;10:2158-67.
49. Deffie AM, McPherson JP, Gupta RS, Hedley DW, Goldenberg GJ. Multifactorial resistance to antineoplastic agents in drug-resistant P388 murine leukemia, Chinese hamster ovary, and human HeLa cells, with emphasis on the role of DNA topoisomerase II. Biochem Cell Biol 1992;70:354-64.
50. Friche E, Danks MK, Schmidt CA, Beck WT. Decreased DNA topoisomerase II in daunorubicin-resistant Ehrlich ascites tumor cells. Cancer Res 1991;51:4213-8.
51. Jordan MA, Wilson L. Microtubules as a target for anticancer drugs. Nat Rev Cancer 2004;4:253-65.
52. Jordan MA, Wendell K, Gardiner S, Derry WB, Copp H, et al. Mitotic block induced in HeLa cells by low concentrations of paclitaxel (Taxol) results in abnormal mitotic exit and apoptotic cell death. Cancer Res 1996;56:816-25.
53. Jordan MA, Toso RJ, Thrower D, Wilson L. Mechanism of mitotic block and inhibition of cell proliferation by taxol at low concentrations. Proc Natl Acad Sci U S A 1993;90:9552-6.
54. Burkhart CA, Kavallaris M, Horwitz SB. The role of β-tubulin isotypes in resistance to antimitotic drugs. BBA-Rev Cancer 2001;2:O1-9.
55. Giannakakou P, Sackett DL, Kang YK, Zhan Z, Buters JT, et al. Paclitaxel-resistant human ovarian cancer cells have mutant β-tubulins that exhibit impaired paclitaxel-driven polymerization. J Biol Chem 1997;272:17118-25.
56. Stover EH, Konstantinopoulos PA, Matulonis UA, Swisher EM. Biomarkers of response and resistance to DNA repair targeted therapies. Clin Cancer Res 2016;22:5651-60.
57. Tanaka M, Okazaki T, Suzuki H, Abbruzzese JL, Li D. Association of multi-drug resistance gene polymorphisms with pancreatic cancer outcome. Cancer 2011;117:744-51.
58. Rodrigues AS, Gomes BC, Martins C, Gromicho M, Oliveira NG, et al. DNA repair and resistance to cancer therapy. In: Chen C, editor. New research directions in DNA repair. IntechOpen 2013. Available from: https://www.intechopen.com/books/new-research-directions-in-dna-repair/dna-repair-and-resistance-to-cancer-therapy. [Last accessed on 7 Jan 2019].
59. Faragher AJ, Fry AM. Nek2A kinase stimulates centrosome disjunction and is required for formation of bipolar mitotic spindles. Mol Biol Cell 2003;14:2876-89.
60. Zhou W, Yang Y, Xia J, Wang H, Salama ME, et al. NEK2 induces drug resistance mainly through activation of efflux drug pumps and is associated with poor prognosis in myeloma and other cancers. Cancer Cell 2013;23:48-62.
61. Lord RV, Brabender J, Gandara D, Alberola V, Camps C, et al. Low ERCC1 expression correlates with prolonged survival after cisplatin plus gemcitabine chemotherapy in non-small cell lung cancer. Clin Cancer Res 2002;8:2286-91.
62. Kwon HC, Roh MS, Oh SY, Kim SH, Kim MC, et al. Prognostic value of expression of ERCC1, thymidylate synthase, and glutathione S-transferase P1 for 5-fluorouracil/oxaliplatin chemotherapy in advanced gastric cancer. Ann Oncol 2007;18:504-9.
63. Usanova S, Piée-Staffa A, Sied U, Thomale J, Schneider A, et al. Cisplatin sensitivity of testis tumour cells is due to deficiency in interstrand-crosslink repair and low ERCC1-XPF expression. Mol Cancer 2010;9:1-11.
64. Tazi J, Bakkour N, Stamm S. Alternative splicing and disease. BBA-Mol Basis Dis 2009;1792:14-26.
65. Marani M, Tenev T, Hancock D, Downward J, Lemoine NR. Identification of novel isoforms of the BH3 domain protein Bim which directly activate Bax to trigger apoptosis. Mol Cell Biol 2002;22:3577-89.
66. de Necochea-Campion R, Shouse GP, Zhou Q, Mirshahidi S, Chen CS. Aberrant splicing and drug resistance in AML. J Hematol Oncol 2016;9:1-9.
67. Passiglia F, Listı` A, Castiglia M, Perez A, Rizzo S, et al. EGFR inhibition in NSCLC: New findings . . . . and opened questions? Crit Rev Oncol Hematol 2017;112:126-35.
68. Stark M, Wichman C, Avivi I, Assaraf YG. Aberrant splicing of folylpolyglutamate synthetase as a novel mechanism of antifolate resistance in leukemia. Blood 2009;113:4362-9.
69. Wang Z, Wu Z, Liu Y, Han W. New development in CAR-T cell therapy. J Hematol Oncol 2017;10:1-11.
70. Wang Y, Bernhardy AJ, Cruz C, Krais JJ, Nacson J, et al. The BRCA1-D11q alternative splice isoform bypasses germline mutations and promotes therapeutic resistance to PARP inhibition and cisplatin. Cancer Res 2016;76:2778-90.
71. García-Becerra R, Santos N, Díaz L, Camacho J. Mechanisms of resistance to endocrine therapy in breast cancer: focus on signaling pathways, miRNAs and genetically based resistance. Int J Mol Sci 2012;14:108-45.
72. Ma G, Ren Y, Wang K, He J. SRC-3 has a role in cancer other than as a nuclear receptor coactivator. Int J Biol Sci 2011;7:664-72.
73. Yan J, Tsai SY, Tsai MJ. SRC-3/AIB1: transcriptional coactivator in oncogenesis. Acta Pharmacol Sin 2006;27:387-94.
74. Catalanotto C, Cogoni C, Zardo G. MicroRNA in control of gene expression: an overview of nuclear functions. Int J Mol Sci 2016;17:1-17.
75. Shivdasani RA. MicroRNAs: regulators of gene expression and cell differentiation. Blood 2006;108:3646-53.
76. Jalkanen AL, Coleman SJ, Wilusz J. Determinants and implications of mRNA poly (A) tail size-does this protein make my tail look big? Semin Cell Dev Biol 2014;34:24-32.
77. Liu H, Wu X, Huang J, Peng J, Guo L. miR-7 modulates chemoresistance of small cell lung cancer by repressing MRP 1/ABCC 1. Int J Exp Pathol 2015;96:240-7.
78. Munoz JL, Rodriguez-Cruz V, Ramkissoon SH, Ligon KL, Greco SJ, et al. Temozolomide resistance in glioblastoma occurs by miRNA-9-targeted PTCH1, independent of sonic hedgehog level. Oncotarget 2015;6:1190-201.
79. Fang Y, Xu C, Fu Y. MicroRNA-17-5p induces drug resistance and invasion of ovarian carcinoma cells by targeting PTEN signaling. J Biol Res-Thessalon 2015;22:1-10.
80. Zhu X, Li Y, Xie C, Yin X, Liu Y, et al. miR-145 sensitizes ovarian cancer cells to paclitaxel by targeting Sp1 and Cdk6. Int J Cancer 2014;135:1286-96.
81. Li H, Zhang P, Sun X, Sun Y, Shi C, et al. MicroRNA-181a regulates epithelial-mesenchymal transition by targeting PTEN in drug-resistant lung adenocarcinoma cells. Int J Oncol 2015;47:1379-92.
82. De Mattos-Arruda L, Bottai G, Nuciforo PG, Di Tommaso L, Giovannetti E, et al. MicroRNA-21 links epithelial-to-mesenchymal transition and inflammatory signals to confer resistance to neoadjuvant trastuzumab and chemotherapy in HER2-positive breast cancer patients. Oncotarget 2015;6:37269-80.
83. Wang Z, Wang N, Liu P, Chen Q, Situ H, et al. MicroRNA-25 regulates chemoresistance-associated autophagy in breast cancer cells, a process modulated by the natural autophagy inducer isoliquiritigenin. Oncotarget 2014;5:7013-26.
84. Zhang Y, Qu X, Li C, Fan Y, Che X, et al. miR-103/107 modulates multidrug resistance in human gastric carcinoma by downregulating Cav-1. Tumor Biol 2015;36:2277-85.
85. Lu L, Ju F, Zhao H, Ma X. MicroRNA-134 modulates resistance to doxorubicin in human breast cancer cells by downregulating ABCC1. Biotechnol Lett 2015;37:2387-94.
86. Feng R, Dong L. Knockdown of microRNA-127 reverses adriamycin resistance via cell cycle arrest and apoptosis sensitization in adriamycin-resistant human glioma cells. Int J Clin Exp Pathol 2015;8:6107-16.
87. Wu Q, Yang Z, Xia L, Nie Y, Wu K, et al. Methylation of miR-129-5p CpG island modulates multi-drug resistance in gastric cancer by targeting ABC transporters. Oncotarget 2014;5:11552-63.
88. Shang Y, Zhang Z, Liu Z, Feng B, Ren G, et al. miR-508-5p regulates multidrug resistance of gastric cancer by targeting ABCB1 and ZNRD1. Oncogene 2014;33:3267-76.
89. To KK, Leung WW, Ng SS. Exploiting a novel miR-519c-HuR-ABCG2 regulatory pathway to overcome chemoresistance in colorectal cancer. Exp Cell Res 2015;338:222-31.
90. Li JH, Luo N, Zhong MZ, Xiao ZQ, Wang JX, et al. Inhibition of MicroRNA-196a might reverse cisplatin resistance of A549/DDP non-small-cell lung cancer cell line. Tumor Biol 2016;37:2387-94.
91. van Jaarsveld MT, van Kuijk PF, Boersma AW, Helleman J, van IWF, et al. miR-634 restores drug sensitivity in resistant ovarian cancer cells by targeting the Ras-MAPK pathway. Mol Cancer 2015;14:196.
92. Sui H, Cai GX, Pan SF, Deng WL, Wang YW, et al. miR200c attenuates P-gp-mediated MDR and metastasis by targeting JNK2/c-Jun signaling pathway in colorectal cancer. Mol Cancer Ther 2014;13:3137-51.
93. Shen X, Guo Y, Qi J, Shi W, Wu X, et al. Study on the association between miRNA-202 expression and drug sensitivity in multiple myeloma cells. Pathol Oncol Res 2016;22:531-9.
94. Zhang AX, Lu FQ, Yang YP, Ren XY, Li ZF, et al. MicroRNA-217 overexpression induces drug resistance and invasion of breast cancer cells by targeting PTEN signaling. Cell Biol Int 2015;42:1455.
95. Gullà A, Di Martino MT, Gallo Cantafio ME, Morelli E, Amodio N, et al. A 13 mer LNA-i-miR-221 inhibitor restores drug sensitivity in melphalan-refractory multiple myeloma cells. Clin Cancer Res 2016;22:1222-33.
96. Hiraki M, Nishimura J, Takahashi H, Wu X, Takahashi Y, et al. Concurrent targeting of KRAS and AKT by MiR-4689 is a novel treatment against mutant KRAS colorectal cancer. Mol Ther Nucleic Acids 2015;4:e231.
99. Myatt SS, Lam EW. The emerging roles of forkhead box (Fox) proteins in cancer. Nat Rev Cancer 2007;7:847-59.
100. Lam EW, Francis RE, Petkovic M. FOXO transcription factors: key regulators of cell fate. Biochem Soc Trans 2006;34:722-6.
101. Abounader R. Interactions between PTEN and receptor tyrosine kinase pathways and their implications for glioma therapy. Expert Rev Anticancer Ther 2009;9:235-45.
102. Sunters A, Fernandez de Mattos S, Stahl M, Brosens JJ, Zoumpoulidou G, et al. FoxO3a transcriptional regulation of Bim controls apoptosis in paclitaxel treated breast cancer cell lines. J Biol Chem 2003;278:49795-805.
103. Sunters A, Madureira PA, Pomeranz KM, Aubert M, Brosens JJ, et al. Paclitaxel-induced nuclear translocation of FOXO3a in breast cancer cells is mediated by c-Jun NH2-terminal kinase and Akt. Cancer Res 2006;66:212-20.
104. Hui RC, Francis RE, Guest SK, Costa JR, Gomes AR, et al. Doxorubicin activates FOXO3a to induce the expression of multidrug resistance gene ABCB1 (MDR1) in K562 leukemic cells. Mol Cancer Ther 2008;7:670-8.
105. Hui RC, Gomes AR, Constantinidou D, Costa JR, Karadedou CT, et al. The forkhead transcription factor FOXO3a increases phosphoinositide-3 kinase/ Akt activity in drug-resistant leukemic cells through induction of PIK3CA expression. Mol Cell Biol 2008;28:5886-98.
106. Saba R, Alsayed A, Zacny JP, Dudek AZ. The role of forkhead box protein m1 in breast cancer progression and resistance to therapy. Int J Breast Cancer 2016;2016:1-8.
107. Tan Y, Raychaudhuri P, Costa RH. Chk2 mediates stabilization of the FoxM1 transcription factor to stimulate expression of DNA repair genes. Mol Cell Biol 2007;27:1007-16.
108. Park YY, Jung SY, Jennings NB, Rodriguez-Aguayo C, Peng G, et al. FOXM1 mediates Dox resistance in breast cancer by enhancing DNA repair. Carcinogenesis 2012;33:1843-53.
109. Monteiro LJ, Khongkow P, Kongsema M, Morris JR, Man C, et al. The Forkhead Box M1 protein regulates BRIP1 expression and DNA damage repair in epirubicin treatment. Oncogene 2013;32:4634-45.
110. M Golubovskaya V. Focal adhesion kinase as a cancer therapy target. Anticancer Agents Med Chem 2010;10:735-41.
111. van Nimwegen MJ, van de Water B. Focal adhesion kinase: a potential target in cancer therapy. Biochem pharmacol 2007;73:597-609.
112. Ren XD, Kiosses WB, Sieg DJ, Otey CA, Schlaepfer DD, et al. Focal adhesion kinase suppresses Rho activity to promote focal adhesion turnover. J Cell Sci 2000;113:3673-8.
113. Roy-Luzarraga M, Hodivala-Dilke K. Molecular pathways: Endothelial cell FAK-a target for cancer treatment. Clin Cancer Res 2016;22:3718-24.
114. Gilbert LA, Hemann MT. DNA damage-mediated induction of a chemoresistant niche. Cell 2010;143:355-66.
115. Lokman NA, Ween MP, Oehler MK, Ricciardelli C. The role of annexin A2 in tumorigenesis and cancer progression. Cancer Microenviron 2011;4:199-208.
116. Grindheim AK, Saraste J, Vedeler A. Protein phosphorylation and its role in the regulation of Annexin A2 function. Biochim Biophys Acta Gen Subj 2017;1861:2515-29.
117. Xu XH, Pan W, Kang LH, Feng H, Song YQ. Association of annexin A2 with cancer development. Oncol rep 2015;33:2121-8.
118. Spijkers-Hagelstein JA, Pinhancos SM, Schneider P, Pieters R, Stam RW. Src kinase-induced phosphorylation of annexin A2 mediates glucocorticoid resistance in MLL-rearranged infant acute lymphoblastic leukemia. Leukemia 2013;27:1063-71.
119. Chen CY, Lin YS, Chen CH, Chen YJ. Annexin A2-mediated cancer progression and therapeutic resistance in nasopharyngeal carcinoma. J Biomed Sci 2018;25:1-10.
120. Liu X, Wei L, Zhao B, Cai X, Dong C, et al. Low expression of KCNN3 may affect drug resistance in ovarian cancer. Mol Med Rep 2018;18:1377-86.
121. Ong HL, Cheng KT, Liu X, Bandyopadhyay BC, Paria BC, et al. Dynamic assembly of TRPC1-STIM1-Orai1 ternary complex is involved in store-operated calcium influx Evidence for similarities in store-operated and calcium release-activated calcium channel components. J Biol Chem 2007;282:9105-16.
122. Dasgupta S, Wasson LM, Rauniyar N, Prokai L, Borejdo J, et al. Novel gene C17orf37 in 17q12 amplicon promotes migration and invasion of prostate cancer cells. Oncogene 2009;28:2860-72.
123. Rajendiran S, Parwani AV, Hare RJ, Dasgupta S, Roby RK, et al. MicroRNA-940 suppresses prostate cancer migration and invasion by regulating MIEN1. Mol cancer 2014;13:1-15.
124. Leung TH, Wong SC, Chan KK, Chan DW, Cheung AN, et al. The interaction between C35 and ΔNp73 promotes chemo-resistance in ovarian cancer cells. Br J Cancer 2013;109:965-75.
125. Wang Y, Chen K, Cai Y, Cai Y, Yuan X, et al. Annexin A2 could enhance multidrug resistance by regulating NF-κB signaling pathway in pediatric neuroblastoma. J Exp Clin Cancer Res 2017;36:1-16.
126. Liu S, Yin F, Zhao M, Zhou C, Ren J, et al. The homing and inhibiting effects of hNSCs-BMP4 on human glioma stem cells. Oncotarget 2016;7:17920-31.
127. Maziveyi M, Alahari SK. Cell matrix adhesions in cancer: the proteins that form the glue. Oncotarget 2017;8:48471-87.
128. Annunziata CM, Kohn EC. Novel facts about FAK: new connections to drug resistance? J Natl Cancer Inst 2013;105:1430-1.
129. Barreiro-Alonso A, Lamas-Maceiras M, García-Díaz R, Rodríguez-Belmonte E, Yu L, et al. Delineating the HMGB1 and HMGB2 interactome in prostate and ovary epithelial cells and its relationship with cancer. Oncotarget 2018;9:19050-64.
130. Syed N, Chavan S, Sahasrabuddhe NA, Renuse S, Sathe G, et al. Silencing of high-mobility group box 2 (HMGB2) modulates cisplatin and 5-fluorouracil sensitivity in head and neck squamous cell carcinoma. Proteomics 2015;15:383-93.
131. Ukmar G, Melloni GE, Raddrizzani L, Rossi P, Di Bella S, et al. PATRI, a Genomics Data Integration Tool for Biomarker Discovery. BioMed Res Int 2018;2018:1-13.
132. Katz E, Dubois-Marshall S, Sims AH, Faratian D, Li J, et al. A gene on the HER2 amplicon, C35, is an oncogene in breast cancer whose actions are prevented by inhibition of Syk. Br J Cancer 2010;103:401-10.
134. Saddoughi SA, Song P, Ogretmen B. Roles of bioactive sphingolipids in cancer biology and therapeutics. In Lipids in Health and Disease. Dordrecht: Springer; 2008. pp. 413-40.
135. Hinrichs JW, Klappe K, Kok JW. Rafts as missing link between multidrug resistance and sphingolipid metabolism. J Membr Biol 2005;203:57-64.
136. Gouaze-Andersson V, Cabot MC. Glycosphingolipids and drug resistance. Biochim Biophys Acta 2006;1758:2096-103.
137. Gouaze V, Liu YY, Prickett CS, Yu JY, Giuliano AE, et al. Glucosylceramide synthase blockade down-regulates P-glycoprotein and resensitizes multidrug-resistant breast cancer cells to anticancer drugs. Cancer Res 2005;65:3861-7.
138. Gouaze-Andersson V, Yu JY, Kreitenberg AJ, Bielawska A, Giuliano AE, et al. Ceramide and glucosylceramide upregulate expression of the multidrug resistance gene MDR1 in cancer cells. Biochim Biophys Acta 2007;1771:1407-17.
139. Akao Y, Banno Y, Nakagawa Y, Hasegawa N, Kim TJ, et al. High expression of sphingosine kinase 1 and S1P receptors in chemotherapy-resistant prostate cancer PC3 cells and their camptothecin-induced up-regulation. Biochem Biophys Res Commun 2006;342:1284-90.
140. Alexander S, Min J, Alexander H. Dictyostelium discoideum to human cells: pharmacogenetic studies demonstrate a role for sphingolipids in chemoresistance. Biochim Biophys Acta 2006;1760:301-9.
141. Baran Y, Salas A, Senkal CE, Gunduz U, Bielawski J, et al. Alterations of ceramide/sphingosine 1-phosphate rheostat involved in the regulation of resistance to imatinib-induced apoptosis in K562 human chronic myeloid leukemia cells. J Biol Chem 2007;282:10922-34.
Cite This Article
Export citation file: BibTeX | RIS
OAE Style
Kumar S, Kushwaha PP, Gupta S. Emerging targets in cancer drug resistance. Cancer Drug Resist 2019;2:161-77. http://dx.doi.org/10.20517/cdr.2018.27
AMA Style
Kumar S, Kushwaha PP, Gupta S. Emerging targets in cancer drug resistance. Cancer Drug Resistance. 2019; 2(2): 161-77. http://dx.doi.org/10.20517/cdr.2018.27
Chicago/Turabian Style
Kumar, Shashank, Prem Prakash Kushwaha, Sanjay Gupta. 2019. "Emerging targets in cancer drug resistance" Cancer Drug Resistance. 2, no.2: 161-77. http://dx.doi.org/10.20517/cdr.2018.27
ACS Style
Kumar, S.; Kushwaha PP.; Gupta S. Emerging targets in cancer drug resistance. Cancer Drug Resist. 2019, 2, 161-77. http://dx.doi.org/10.20517/cdr.2018.27
About This Article
Special Issue
Copyright
Data & Comments
Data
 Cite This Article 24 clicks
Cite This Article 24 clicks














Comments
Comments must be written in English. Spam, offensive content, impersonation, and private information will not be permitted. If any comment is reported and identified as inappropriate content by OAE staff, the comment will be removed without notice. If you have any queries or need any help, please contact us at support@oaepublish.com.